
In the past decade, high-throughput technologies produced a huge amount of biological data. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.Ĭompeting interests: The authors have declared that no competing interests exist. CUP B15G13000010006 awarded by the Regione Valle dAosta for the project: “Open Health Care Network Analysis” and by the Italian Ministry of Education, University & Research (MIUR) (Project PRIN 2010, MIND). įunding: This work has been partially supported by Grant No.
CYTOSCAPE ONLINE FULL
ReNE can be freely installed from the Cytoscape App Store ( ) and the full source code is freely available for download through a SVN repository accessible at. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.ĭata Availability: The authors confirm that all data underlying the findings are fully available without restriction. Received: JAccepted: DecemPublished: December 26, 2014Ĭopyright: © 2014 Politano et al. PLoS ONE 9(12):Įditor: Julio Vera, University of Erlangen-Nuremberg, Germany The reliability of the introduced interactions only depends on the reliability of the source data, which is out of control of ReNe developers.Ĭitation: Politano G, Benso A, Savino A, Di Carlo S (2014) ReNE: A Cytoscape Plugin for Regulatory Network Enhancement. ReNE enhances a network by only integrating data from public repositories, without any inference or prediction. The enhanced network produced by ReNE is exportable in multiple formats for further analysis via third party applications. The resulting enhanced network is still a fully functional Cytoscape network where each regulatory element (transcription factor, miRNA, gene, protein) and regulatory mechanism (up-regulation/down-regulation) is clearly visually identifiable, thus enabling a better visual understanding of its role and the effect in the network behavior. The normalized network is then analyzed to include missing transcription factors, miRNAs, and proteins.
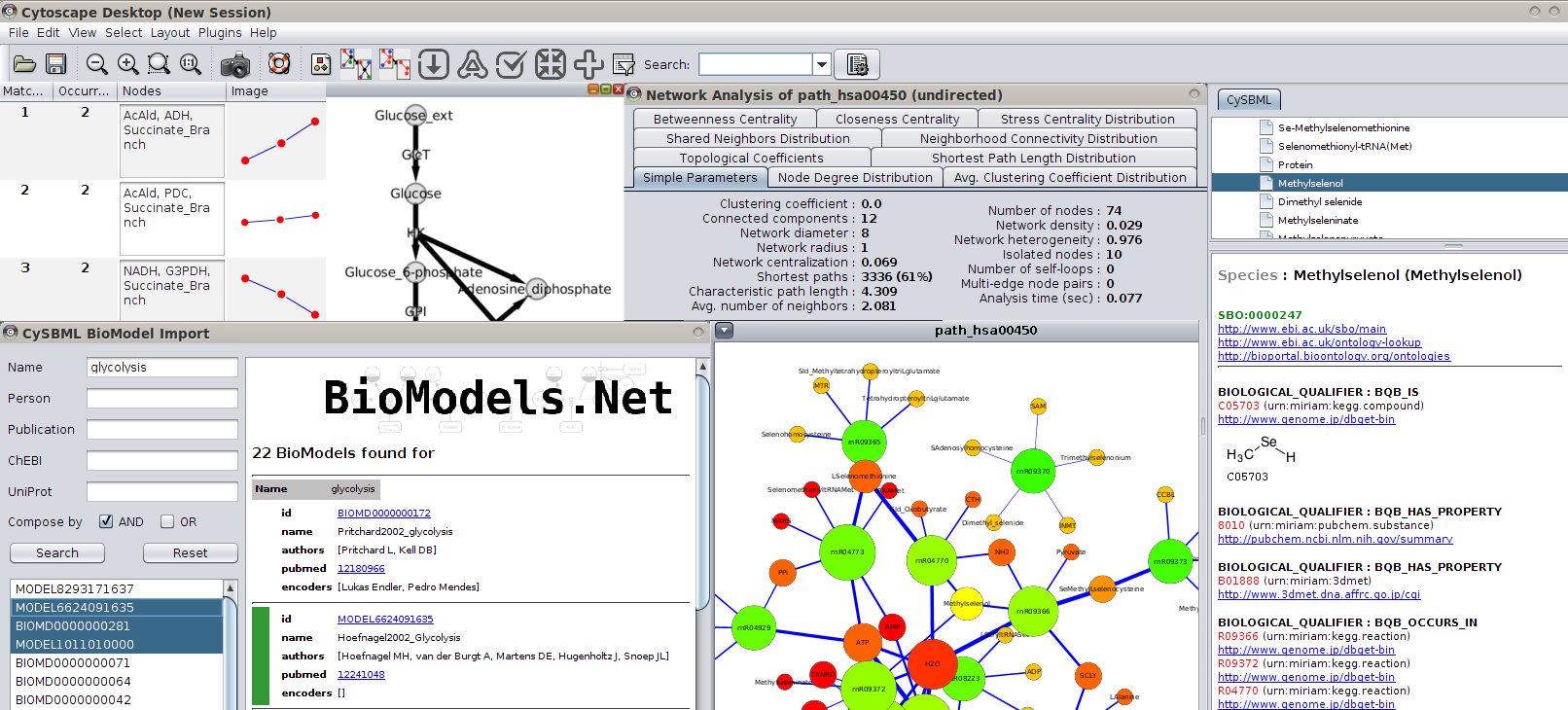

The merged network structure is normalized to guarantee a consistent and uniform description of the network nodes and edges and to enrich all integrated data with additional annotations retrieved from genome-wide databases like NCBI, thus producing a pathway fully manageable through the Cytoscape environment. Moreover, ReNE allows researchers to merge multiple pathways coming from different sources. ReNE can automatically import a network layout from the Reactome or KEGG repositories, or work with custom pathways described using a standard OWL/XML data format that the Cytoscape import procedure accepts. In this context, we introduce ReNE, a Cytoscape 3.x plugin designed to automatically enrich a standard gene-based regulatory network with more detailed transcriptional, post transcriptional, and translational data, resulting in an enhanced network that more precisely models the actual biological regulatory mechanisms. To integrate networks with post transcriptional regulatory data, researchers are therefore forced to manually resort to specific third party databases. Regulatory networks are commonly limited to gene entities. Despite post transcriptional regulatory elements (i.e., miRNAs) are widely investigated in current research, their usage and visualization in biological networks is very limited. One of the biggest challenges in the study of biological regulatory mechanisms is the integration, americanmodeling, and analysis of the complex interactions which take place in biological networks.


 0 kommentar(er)
0 kommentar(er)
